Artificial Intelligence Shaping the Next Generation of Viruses
With each passing day, researchers continue to find novel applications of artificial intelligence (AI). Many people know of the power of large language model chatbots like ChatGPT, but AI can also decode a different kind of language: the genetic code of living organisms and viruses. Most recently, researchers at the Hie Lab at Stanford University and the Arc Institute successfully utilized AI to design viruses that are able to hunt and kill strains of E. Coli bacteria. Although this study by Samuel King and colleagues is yet to be peer reviewed, the authors believe that this development, in conjunction with existing therapeutic treatments, holds the potential to treat bacterial infections (1).
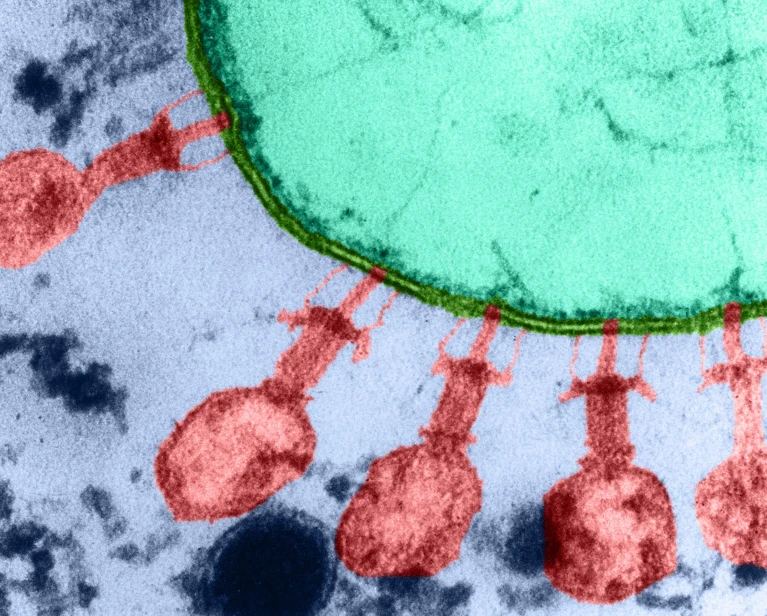
While the idea of an AI generating a functioning virus seems like science fiction, the process is grounded in biology and computation. The model the scientists used, Evo 2, pored over 128,000 genomes of humans, plants, and other eukaryotic organisms—a large step from the previous model which only trained on 80,000 bacteria, archaea, and viruses (2). This leap from the shorter, circular, genomes of prokaryotes (bacteria and archaea) to the longer, linear, sequences of eukaryotic genomes represents a significant jump in complexity. Eukaryotes have genes interspersed with many non-coding regions, or introns, that regulate the transcription of expressed genes, exons. Particularly, these introns can be thousands of base pairs long and far from the genes they control (3). According to one of the computational genomicists at Stanford, the Evo 2 model, with its vast training data set, is able to better recognize patterns that occur in exons and predict whether mutations would cause disease (2). There is, however, some uncertainty regarding how well Evo 2 understands the impact of distant introns (2).
Though, the most important outcome of AI advancement in this field is the new ability to generate DNA sequences for specific purposes (4). King and his team focused their efforts on generating a bacteriophage, a virus that infects and replicates within bacteria, capable of targeting specific strains of E. coli. Their team used a well-studied bacteriophage named ΦX174 as the template genome, training Evo 1 and 2 on the phage’s genome to better apply constraints and filters as sequences are being generated to result in the desired targeting characteristics (4).
16 of the 300 generated genomes were found to have tropism—a viruses’ preference for a specific kind of cell—successfully limited to E. coli bacteria (4). Furthermore, some of these phages possessed genetic differences that allowed them to reproduce quicker and outcompete ΦX174 after only six hours (4).
King’s team then assessed the efficacy of these generated phages against E. coli strains resistant to ΦX174. They created a combination of all 16 generated phages at a similar concentration to a “ΦX174-only cocktail,” and compared their efficacy against three ΦX174 resistant E. coli strains. Their results were quite striking as the generated phage cocktail was able to successfully inhibit the growth of all three strains, whereas the ΦX174-only cocktail could not inhibit any (4). Further analysis of the phage genomes responsible for overcoming each strain’s resistance showed that the success likely came from recombinations and mutations of the AI generated phages (4).
Though there is great potential to ameliorate lives of people across the world through this technology, at the same time, there are biosafety concerns of people using this technology to generate viruses that could harm humans. Though the team has taken precaution to exclude “viruses that affect eukaryotes, including humans, from the Evo models’ training data,” these ethical questions are always worth keeping in mind (1).
Sources:
- Kavanagh, K. (2025). World’s first AI-designed viruses a step towards AI-generated life. Nature. https://doi.org/10.1038/d41586-025-03055-y
- Callaway, E. (2025). Biggest-ever AI biology model writes DNA on demand. Nature. https://doi.org/10.1038/d41586-025-00531-3
- Long, M., & Deutsch, M. (1999). Intron—exon structures of eukaryotic model organisms. Nucleic Acids Research, 27(15), 3219–3228. https://doi.org/10.1093/nar/27.15.3219
- King, S. H., Driscoll, C. L., Li, D. B., Guo, D., Merchant, A. T., Garyk Brixi, Wilkinson, M. E., & Hie, B. L. (2025). Generative design of novel bacteriophages with genome language models. BioRxiv (Cold Spring Harbor Laboratory). https://doi.org/10.1101/2025.09.12.67591
Image:


Comments are closed.