A CRISPR-based paper test strip’s potential to redefine influenza testing
Each year, influenza afflicts one billion people worldwide, with approximately forty million cases concentrated in the United States, and contributes to more than 300,000 mortalities (1). With countless symptoms including headaches, body aches, sore throats, and fever, this contagious virus forces the masses to rely on over-the-counter medicine and prescriptions like oseltamivir, commonly known as Tamiflu (2). A crucial part of improving the rate of infection and overall outbreak response is enhancing diagnostic testing. As of right now, the gold standard for influenza diagnosis is the quantitative reverse transcription-polymerase chain reaction (RT-PCR) process that yields accurate results with high sensitivity (3). However, fewer than 1% of people who eventually contract the virus get tested, largely due to expensive prices; most RT-PCR tests require trained operators and costly equipment (4). Yet Cambridge’s Broad Institute, in collaboration with the Massachusetts Institute of Technology, Harvard University, and Princeton University, unlocked a powerful alternative to RT-PCR tests for influenza in July 2024: rapid nucleic acid paper tests that specify influenza type and strain (4).
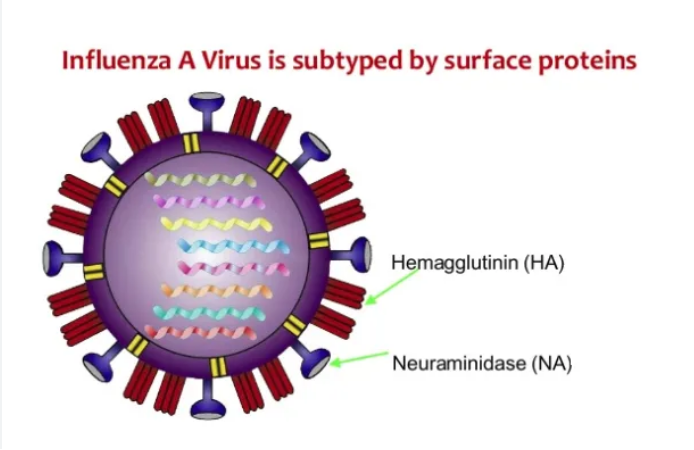
While both influenza A and B contribute to seasonal epidemics, scientists have identified more than 130 influenza A combinations (5). Classified by two surface proteins, hemagglutinin (H) and neuraminidase (N), influenza A has 18 different H subtypes and 11 N subtypes (5). The H and N proteins act as antigens, triggering the host’s immune response (5). But if the virus’ antigens are different from those contained in flu vaccines, the vaccine will not neutralize the body’s immune response (5). The Broad Institute’s simple flu test distinguishes between influenza A, influenza B, and the prevalent influenza A H1N1 and H3N2 variants (4). Additionally, it detects strains that do not neutralize with oseltamivir, which can eventually help clinicians prescribe more appropriate antivirals (4, 7).
Influenza A’s two surface proteins, hemagglutinin and neuraminidase (2).
Cognizant of the need for diagnostic technologies that combine precision and speed, the Broad Institute utilized machine learning softwares and clustered regularly interspaced short palindromic repeats (CRISPR) technologies. Using the Streamlined Highlighting of Infections to Navigate Epidemics (SHINE) method to detect viral RNA, researchers utilized CRISPR enzymes to detect these genetic sequences found in samples (6). Four distinct SHINE assays were developed to differentiate between influenza A, its H1N1 and H3N2 variants, and influenza B, and to identify mutations (6). Data obtained from the National Center for Biotechnology Information Influenza Database from 2016 to 2021 allowed the researchers to highlight relevant strains of influenza (7). This information contributed to the designing of the influenza assays, in which ADAPT, a machine learning-based software, identified CRISPR RNA (crRNA) molecules. crRNA molecules contain the viral DNA which guides the CRISPR proteins to the sequence at which the cutting molecule splits the DNA (8).
Similarly, ADAPT determines primer pairs, which hybridize with influenza’s genome to amplify a specific region, in order to capture the variant of influenza in a comprehensive manner (7). As additional components of the assay’s reagents, Reverse Transcription Recombinase Polymerase Amplification (RT-RPA) amplifies the viral genome and works in tandem with the programmed Cas13 protein to target the RNA sequences of influenza from various nasopharyngeal samples (7). These actions follow the splicing of viral RNA with the LwaCas13a and EnGen Lba Cas12a proteins (7). As the various constituents bind to the target DNA, signals from two fluorescent dyes, FAM and HEX, are released (7). Researchers discovered positive tests for influenza produced two blue-green lines as a result of the fluorescence––one at the top of the paper strip and another at the bottom––while negative tests only maintain a visible line at the bottom (7).
A graphic of the CRISPR-based paper strip test process (1).
With a foundational model for the paper test strips, the scientists aimed to simplify this CRISPR-based diagnostic test. Freeze-drying the reagents produced pellets, which can be rehydrated and aid with transport and storage in the long-term (7). In addition, the researchers leveraged the assays by adding an internal control to verify their integrity. In this case, human RNase P carries out the cleavage reaction and targets viral RNA down to 1000 copies/µL (7). Further side-by-side testing of this diagnostic method with RT-PCR tests revealed an impressive 100% compliance for five influenza A viral samples (two H1N1 and three H3N2 strains), one influenza B sample, and ten influenza-negative samples (7).
Dr. Cameron Myhrvold, a key collaborator on this project from Princeton University, responded to the successful and revolutionary results. “We hope these tests will be as simple as rapid antigen tests, and they’ll still have the specificity and performance of a nucleic acid test that would normally be done in a laboratory setting,” he wrote (6). Distributing these tests to the public will not be the only advantage of this notable research from the Broad Institute. The enzymatic, CRISPR-based method of SHINE that delineates genetic sequences found in viral RNA is adaptive (3). Therefore, scientists across the world can take advantage of a similar model to create future at-home tests for other viruses (7).
A highly contagious respiratory virus, influenza spreads rapidly, especially during annual, seasonal epidemics. The virus’ adaptability and several combinations of defining surface proteins, along with rapid adaptations to their environments through genetic mutations, cause increased proliferation of this disease and reduced societal immunity. As a result, individuals and communities across the world will benefit from accessible testing, like that modeled by the Broad Institute’s groundbreaking prototype. The scientific community must not underestimate the power of paper, especially when combined with CRISPR technologies, machine learning methods, and the collaboration of several institutions.
Sources:
- World Health Organization. (2024, March 30). The Burden of Influenza. World Health Organization. Retrieved from. https://www.who.int/news-room/feature-stories/detail/the-burden-of-influenza.
- (2023, September 4). Flu (Influenza): Causes, Symptoms, Types & Treatment. Cleveland Clinic. Retrieved from https://my.clevelandclinic.org/health/diseases/4335-influenza-flu.
- CDC. (2024, August 19). Rapid Diagnostic Testing for Influenza: Information for Clinical Laboratory Directors. Influenza (Flu). Center for Disease Control and Prevention. Retrieved from https://www.cdc.gov/flu/php/laboratories/rapidlab.html.
- (2024, June 18). Simple test for flu could improve diagnosis and surveillance. Broad Institute. Retrieved from https://www.broadinstitute.org/news/simple-test-flu-could-improve-diagnosis-and-surveillance.
- CDC. (2024, September 27). Types of Influenza Viruses. Influenza (Flu). Retrieved from https://www.cdc.gov/flu/about/viruses-types.html.
- Van Beuskeom, Mary. (2024, June 24). Paper strip test can identify flu subtypes, may have other applications, scientists say. CIDRAP. Retrieved from https://www.cidrap.umn.edu/influenza-variants/paper-strip-test-can-identify-flu-subtypes-may-have-other-applications.
- Zhang, Yibin B. et al. (2024 July). CRISPR-Based Assays for Point-of-Need Detection and Subtyping of Influenza. The Journal of Molecular Diagnostics, Volume 26, Issue 7, 599 – 612. Retrieved from https://www.jmdjournal.org/article/S1525-1578(24)00087-4/fulltext#sec-1-6.
- (2024). Functioning of CRISPR-Cas9. Retrieved from https://www.mpg.de/11824433/crispr-cas9-functioning.
Images:






Comments are closed.